Side Menu
BRAF
Basic information
- Official gene symbol
-
B-Raf proto-oncogene, serine/threonine kinase
- Conventional gene symbol
-
B-RAF1, B-raf, BRAF1, NS7, RAFB1
- Classification
-
Oncogene
- Official gene ID
- Transcript ID
- Functional classification
-
Tumor growth and progression
- Signaling pathway
-
MAPK
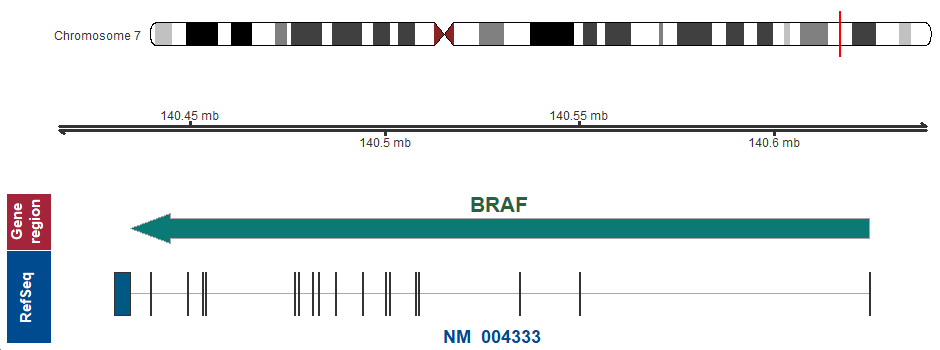
- Chromosomal location
-
7q34 (chr7:140434397..140624503, complement)
- Protein length
-
766
Chromosome ideogram
Pathway map

Frequency of somatic alterationsTumor type
Base substitutions, insertions, and deletions

Copy number alterations

Distribution of tumor mutation burdenTMB

List of driver somatic mutations
| Genomic Coordinates (GRCh37/hg19) | Reference | Variant | Exon | Amino Acid change | Coding DNA change | COSMIC (v92) | Classification | No. of Samples |
|---|---|---|---|---|---|---|---|---|
| chr7:140501331 | A | C | 6 / 18 | p.F247L | c.741T>G | COSV56165267 | Tier2 | 1 / 200 |
| chr7:140481393 | T | C | 11 / 18 | p.Y472C | c.1415A>G | COSV56067953 | Tier1 | 1 / 200 |
| chr7:140481397 | C | A | 11 / 18 | p.V471F | c.1411G>T | COSV56229839 | Tier1 | 1 / 200 |
| chr7:140481402 | C | G | 11 / 18 | p.G469A | c.1406G>C | COSV56061424 | Tier1 | 4 / 200 |
| chr7:140481402 | C | T | 11 / 18 | p.G469E | c.1406G>A | COSV56065622 | Tier1 | 2 / 200 |
| chr7:140481403 | C | T | 11 / 18 | p.G469R | c.1405G>A | COSV56070557 | Tier1 | 1 / 200 |
| chr7:140481403 | C | G | 11 / 18 | p.G469R | c.1405G>C | COSV56082352 | Tier1 | 1 / 200 |
| chr7:140481408 | G | A | 11 / 18 | p.S467L | c.1400C>T | COSV56106484 | Tier1 | 1 / 200 |
| chr7:140481411 | C | T | 11 / 18 | p.G466E | c.1397G>A | COSV56059390 | Tier1 | 1 / 200 |
| chr7:140481411 | C | G | 11 / 18 | p.G466A | c.1397G>C | COSV56070593 | Tier1 | 1 / 200 |
| chr7:140481411 | C | A | 11 / 18 | p.G466V | c.1397G>T | COSV56057462 | Tier1 | 2 / 200 |
| chr7:140481412 | C | G | 11 / 18 | p.G466R | c.1396G>C | COSV56064038 | Tier2 | 1 / 200 |
| chr7:140481412 | C | T | 11 / 18 | p.G466R | c.1396G>A | COSV56099462 | Tier2 | 1 / 200 |
| chr7:140481478 | G | A | 11 / 18 | p.R444W | c.1330C>T | COSV56083156 | Tier2 | 1 / 200 |
| chr7:140477836 | GTAGGTGCTGTCACAT | G | 12 / 18 | p.N486_P490del | c.1457_1471delATGTGACAGCACCTA | COSV56100024 | Tier2 | 1 / 200 |
| chr7:140477853 | C | G | 12 / 18 | p.L485F | c.1455G>C | Tier2 | 1 / 200 | |
| chr7:140477861 | T | C | 12 / 18 | p.K483E | c.1447A>G | COSV56236813 | Tier2 | 1 / 200 |
| chr7:140453132 | T | A | 15 / 18 | p.K601N | c.1803A>T | COSV56058093 | Tier1 | 1 / 200 |
| chr7:140453134 | T | C | 15 / 18 | p.K601E | c.1801A>G | COSV56058494 | Tier1 | 6 / 200 |
| chr7:140453136 | A | T | 15 / 18 | p.V600E | c.1799T>A | COSV56056643 | Tier1 | 100 / 200 |
| chr7:140453145 | A | T | 15 / 18 | p.L597Q | c.1790T>A | COSV56072463 | Tier1 | 1 / 200 |
| chr7:140453148 | C | A | 15 / 18 | p.G596V | c.1787G>T | COSV99951808 | Tier2 | 2 / 200 |
| chr7:140453154 | T | C | 15 / 18 | p.D594G | c.1781A>G | COSV56065695 | Tier1 | 10 / 200 |
| chr7:140453154 | T | G | 15 / 18 | p.D594A | c.1781A>C | COSV56224199 | Tier1 | 1 / 200 |
| chr7:140453155 | C | T | 15 / 18 | p.D594N | c.1780G>A | COSV56061673 | Tier1 | 5 / 200 |
| chr7:140453193 | T | A | 15 / 18 | p.N581I | c.1742A>T | COSV56106422 | Tier1 | 2 / 200 |
| chr7:140453193 | T | C | 15 / 18 | p.N581S | c.1742A>G | COSV56058108 | Tier1 | 2 / 200 |

