Side Menu
BRCA2
Basic information
- Official gene symbol
-
BRCA2 DNA repair associated
- Conventional gene symbol
-
BRCC2, BROVCA2, FACD, FAD, FAD1, FANCD, FANCD1, GLM3, PNCA2, XRCC11
- Classification
-
Tumor suppressor gene
- Official gene ID
- Transcript ID
- Functional classification
-
Genome maintenance
- Signaling pathway
-
Core DNA Damage Response
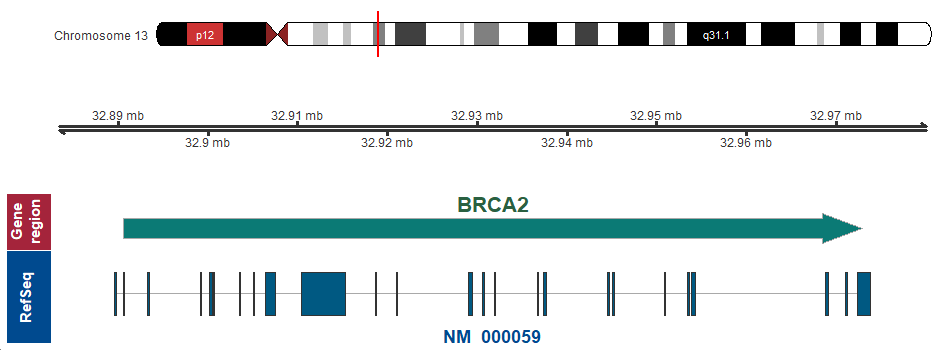
- Chromosomal location
-
13q13.1 (chr13:32890598..32972907)
- Protein length
-
3,418
Chromosome ideogram
Pathway map

Frequency of somatic alterationsTumor type
Base substitutions, insertions, and deletions

Copy number alterations

Distribution of tumor mutation burdenTMB

List of driver somatic mutations
| Genomic Coordinates (GRCh37/hg19) | Reference | Variant | Exon | Amino Acid change | Coding DNA change | COSMIC (v92) | Classification | No. of Samples |
|---|---|---|---|---|---|---|---|---|
| chr13:32890632 | TTGAAATTTTTAAGACACGCTGCA | T | 2 / 27 | p.F12fs | c.36_58delTGAAATTTTTAAGACACGCTGCA | Tier2 | 1 / 156 | |
| chr13:32890633 | TGAAATTTTTAAGACACGCTGCA | T | 2 / 27 | p.E13fs | c.37_58delGAAATTTTTAAGACACGCTGCA | Tier2 | 1 / 156 | |
| chr13:32893291 | G | T | 3 / 27 | p.E49* | c.145G>T | Tier2 | 1 / 156 | |
| chr13:32893393 | G | T | 3 / 27 | p.E83* | c.247G>T | Tier2 | 1 / 156 | |
| chr13:32893435 | G | T | 3 / 27 | p.E97* | c.289G>T | COSV66461936 | Tier2 | 1 / 156 |
| chr13:32906673 | C | A | 10 / 27 | p.S353* | c.1058C>A | COSV101204193 | Tier2 | 1 / 156 |
| chr13:32906996 | G | T | 10 / 27 | p.E461* | c.1381G>T | Tier2 | 1 / 156 | |
| chr13:32910437 | C | T | 11 / 27 | p.Q649* | c.1945C>T | Tier2 | 1 / 156 | |
| chr13:32912453 | GA | G | 11 / 27 | p.D1321fs | c.3962delA | Tier2 | 1 / 156 | |
| chr13:32912858 | G | T | 11 / 27 | p.E1456* | c.4366G>T | Tier2 | 1 / 156 | |
| chr13:32913176 | C | T | 11 / 27 | p.Q1562* | c.4684C>T | COSV101204461 | Tier2 | 1 / 156 |
| chr13:32913593 | C | T | 11 / 27 | p.Q1701* | c.5101C>T | Tier2 | 1 / 156 | |
| chr13:32913651 | C | A | 11 / 27 | p.S1720* | c.5159C>A | Tier2 | 1 / 156 | |
| chr13:32914056 | C | G | 11 / 27 | p.S1855* | c.5564C>G | Tier2 | 1 / 156 | |
| chr13:32914800 | C | G | 11 / 27 | p.S2103* | c.6308C>G | Tier2 | 1 / 156 | |
| chr13:32914877 | G | T | 11 / 27 | p.E2129* | c.6385G>T | COSV66449661 | Tier2 | 2 / 156 |
| chr13:32915135 | TACTC | T | 11 / 27 | p.Y2215fs | c.6644_6647delACTC | Tier2 | 1 / 156 | |
| chr13:32915264 | G | T | 11 / 27 | p.E2258* | c.6772G>T | Tier2 | 1 / 156 | |
| chr13:32931937 | CTT | C | 16 / 27 | p.F2560fs | c.7679_7680delTT | Tier2 | 1 / 156 | |
| chr13:32932019 | G | A | 16 / 27 | p.W2586* | c.7758G>A | COSV66462892 | Tier2 | 1 / 156 |
| chr13:32968936 | AGCAAC | A | 25 / 27 | p.S3123fs | c.9368_9372delGCAAC | Tier2 | 1 / 156 | |
| chr13:32968967 | C | G | 25 / 27 | p.S3133* | c.9398C>G | Tier2 | 1 / 156 | |
| chr13:32972663 | C | A | 27 / 27 | p.S3338* | c.10013C>A | Tier2 | 1 / 156 |

