Side Menu
CDKN2A
Basic information
- Official gene symbol
-
cyclin dependent kinase inhibitor 2A
- Conventional gene symbol
-
ARF, CDKN2, CMM2, INK4, INK4A, MLM, MTS-1, MTS1, P14, P14ARF, P16, P16-INK4A, P16INK4, P16INK4A, P19, P19ARF, TP16
- Classification
-
Tumor suppressor gene
- Official gene ID
- Transcript ID
- Functional classification
-
Cell cycle
- Signaling pathway
-
Cell cycle
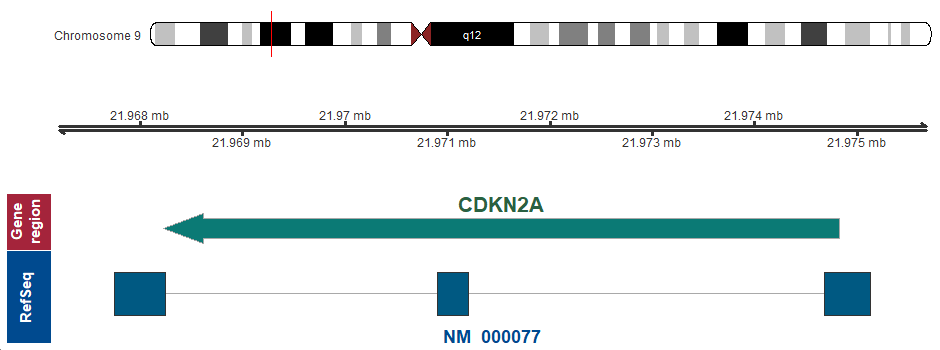
- Chromosomal location
-
9p21.3 (chr9:21968228..21974826, complement)
- Protein length
-
156
Chromosome ideogram
Pathway map

Frequency of somatic alterationsTumor type
Base substitutions, insertions, and deletions

Copy number alterations

Distribution of tumor mutation burdenTMB

List of driver somatic mutations
| Genomic Coordinates (GRCh37/hg19) | Reference | Variant | Exon | Amino Acid change | Coding DNA change | COSMIC (v92) | Classification | No. of Samples |
|---|---|---|---|---|---|---|---|---|
| chr9:21974677 | C | A | 1 / 3 | p.Q50H | c.150G>T | COSV58692109 | Tier2 | 1 / 159 |
| chr9:21974679 | G | A | 1 / 3 | p.Q50* | c.148C>T | COSV58683786 | Tier1 | 1 / 159 |
| chr9:21974684 | G | A | 1 / 3 | p.P48L | c.143C>T | COSV58683273 | Tier1 | 2 / 159 |
| chr9:21974695 | G | C | 1 / 3 | p.Y44* | c.132C>G | COSV58696346 | Tier2 | 2 / 159 |
| chr9:21974730 | C | A | 1 / 3 | p.E33* | c.97G>T | COSV58682729 | Tier2 | 2 / 159 |
| chr9:21974732 | A | G | 1 / 3 | p.L32P | c.95T>C | COSV58685103 | Tier1 | 1 / 159 |
| chr9:21974735 | AGCGCCCGCACCTCCTCTACCCGACCCCGGGCC | A | 1 / 3 | p.R22fs | c.60_91delGGCCCGGGGTCGGGTAGAGGAGGTGCGGGCGC | Tier2 | 1 / 159 | |
| chr9:21974759 | CCCCGGGCCGCGGCCGTGGCCAG | C | 1 / 3 | p.L16fs | c.46_67delCTGGCCACGGCCGCGGCCCGGG | Tier2 | 1 / 159 | |
| chr9:21974776 | GGCCA | G | 1 / 3 | p.L16fs | c.47_50delTGGC | COSV58683946 | Tier2 | 1 / 159 |
| chr9:21974782 | C | T | 1 / 3 | p.W15* | c.45G>A | COSV58683123 | Tier2 | 1 / 159 |
| chr9:21974787 | C | CT | 1 / 3 | p.D14fs | c.39_40insA | Tier2 | 1 / 159 | |
| chr9:21974792 | G | T | 1 / 3 | p.S12* | c.35C>A | COSV58689693 | Tier2 | 1 / 159 |
| chr9:21974799 | C | A | 1 / 3 | p.E10* | c.28G>T | COSV58705987 | Tier2 | 3 / 159 |
| chr9:21970971 | G | T | 2 / 3 | p.Y129* | c.387C>A | COSV58684005 | Tier2 | 2 / 159 |
| chr9:21971000 | C | A | 2 / 3 | p.E120* | c.358G>T | COSV58683444 | Tier2 | 3 / 159 |
| chr9:21971005 | GC | G | 2 / 3 | p.A118fs | c.352delG | Tier2 | 1 / 159 | |
| chr9:21971017 | G | A | 2 / 3 | p.P114L | c.341C>T | COSV58683051 | Tier1 | 2 / 159 |
| chr9:21971028 | C | T | 2 / 3 | p.W110* | c.330G>A | COSV58682827 | Tier2 | 7 / 159 |
| chr9:21971029 | C | T | 2 / 3 | p.W110* | c.329G>A | COSV58682976 | Tier2 | 4 / 159 |
| chr9:21971036 | C | G | 2 / 3 | p.D108H | c.322G>C | COSV58690035 | Tier2 | 2 / 159 |
| chr9:21971036 | C | A | 2 / 3 | p.D108Y | c.322G>T | COSV58682998 | Tier2 | 1 / 159 |
| chr9:21971057 | C | A | 2 / 3 | p.G101W | c.301G>T | COSV58692144 | Tier1 | 1 / 159 |
| chr9:21971068 | A | C | 2 / 3 | p.L97R | c.290T>G | COSV58684740 | Tier1 | 1 / 159 |
| chr9:21971069 | G | GCACC | 2 / 3 | p.L97fs | c.285_288dupGGTG | Tier2 | 1 / 159 | |
| chr9:21971096 | C | A | 2 / 3 | p.E88* | c.262G>T | COSV58683071 | Tier2 | 2 / 159 |
| chr9:21971099 | G | A | 2 / 3 | p.R87W | c.259C>T | COSV58718452 | Tier1 | 2 / 159 |
| chr9:21971108 | C | A | 2 / 3 | p.D84Y | c.250G>T | COSV58683210 | Tier1 | 1 / 159 |
| chr9:21971108 | C | T | 2 / 3 | p.D84N | c.250G>A | COSV58683289 | Tier2 | 1 / 159 |
| chr9:21971110 | T | C | 2 / 3 | p.H83R | c.248A>G | COSV58689512 | Tier2 | 2 / 159 |
| chr9:21971111 | G | A | 2 / 3 | p.H83Y | c.247C>T | COSV58682852 | Tier1 | 6 / 159 |
| chr9:21971111 | G | C | 2 / 3 | p.H83D | c.247C>G | COSV58690009 | Tier2 | 1 / 159 |
| chr9:21971118 | TC | T | 2 / 3 | p.R80fs | c.239delG | COSV58729341 | Tier2 | 1 / 159 |
| chr9:21971120 | G | A | 2 / 3 | p.R80* | c.238C>T | COSV58682746 | Tier2 | 16 / 159 |
| chr9:21971123 | TGA | T | 2 / 3 | p.L78fs | c.233_234delTC | COSV58687173 | Tier2 | 1 / 159 |
| chr9:21971127 | AGTGG | A | 2 / 3 | p.A76fs | c.227_230delCCAC | Tier2 | 1 / 159 | |
| chr9:21971138 | C | T | 2 / 3 | p.D74N | c.220G>A | COSV58688841 | Tier1 | 2 / 159 |
| chr9:21971139 | GGC | G | 2 / 3 | p.A73fs | c.217_218delGC | Tier2 | 1 / 159 | |
| chr9:21971142 | G | T | 2 / 3 | p.C72* | c.216C>A | COSV58683179 | Tier2 | 1 / 159 |
| chr9:21971150 | GC | G | 2 / 3 | p.E69fs | c.207delG | Tier2 | 1 / 159 | |
| chr9:21971150 | GCT | G | 2 / 3 | p.E69fs | c.206_207delAG | Tier2 | 1 / 159 | |
| chr9:21971155 | GC | G | 2 / 3 | p.A68fs | c.202delG | Tier2 | 1 / 159 | |
| chr9:21971174 | GCTCCGCCACT | G | 2 / 3 | p.V59fs | c.174_183delAGTGGCGGAG | Tier2 | 1 / 159 | |
| chr9:21971177 | C | A | 2 / 3 | p.E61* | c.181G>T | COSV58683250 | Tier2 | 4 / 159 |
| chr9:21971179 | GCCACT | G | 2 / 3 | p.V59fs | c.174_178delAGTGG | COSV58693461 | Tier2 | 1 / 159 |
| chr9:21971186 | G | A | 2 / 3 | p.R58* | c.172C>T | COSV58682666 | Tier2 | 14 / 159 |
| chr9:21971197 | A | ATCAT | 2 / 3 | p.M54fs | c.157_160dupATGA | Tier2 | 1 / 159 | |
| chr9:21971198 | T | TCA | 2 / 3 | p.M54fs | c.158_159dupTG | Tier2 | 1 / 159 |

