Side Menu
KDM6A
Basic information
- Official gene symbol
-
lysine demethylase 6A
- Conventional gene symbol
-
KABUK2, UTX, bA386N14.2
- Classification
-
Oncogene / Tumor suppressor gene
- Official gene ID
- Transcript ID
- Functional classification
-
Epigenetic regulation
- Signaling pathway
-
Epigenetic modification
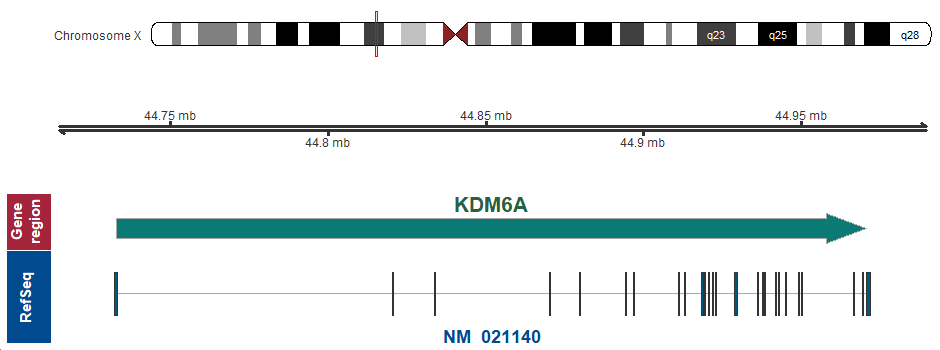
- Chromosomal location
-
Xp11.3 (chrX:44732798..44970656)
- Protein length
-
1,401
Chromosome ideogram
Frequency of somatic alterationsTumor type
Base substitutions, insertions, and deletions

Copy number alterations

Distribution of tumor mutation burdenTMB

List of driver somatic mutations
| Genomic Coordinates (GRCh37/hg19) | Reference | Variant | Exon | Amino Acid change | Coding DNA change | COSMIC (v92) | Classification | No. of Samples |
|---|---|---|---|---|---|---|---|---|
| chrX:44833924 | C | G | 4 / 29 | p.Y116* | c.348C>G | Tier2 | 1 / 117 | |
| chrX:44870264 | G | A | 5 / 29 | p.W148* | c.443G>A | Tier2 | 1 / 117 | |
| chrX:44879904 | C | T | 6 / 29 | p.R165* | c.493C>T | COSV65037080 | Tier2 | 1 / 117 |
| chrX:44896918 | ACT | A | 8 / 29 | p.L214fs | c.639_640delCT | Tier2 | 1 / 117 | |
| chrX:44910995 | GCAGA | G | 9 / 29 | p.T234fs | c.701_704delCAGA | Tier2 | 1 / 117 | |
| chrX:44918502 | C | T | 12 / 29 | p.Q329* | c.985C>T | COSV65045178 | Tier2 | 1 / 117 |
| chrX:44918595 | ACT | A | 12 / 29 | p.Y362fs | c.1083_1084delCT | COSV65040281 | Tier2 | 1 / 117 |
| chrX:44919375 | C | T | 13 / 29 | p.Q435* | c.1303C>T | Tier2 | 1 / 117 | |
| chrX:44922694 | CG | C | 16 / 29 | p.R519fs | c.1556delG | COSV65039445 | Tier2 | 1 / 117 |
| chrX:44922760 | C | T | 16 / 29 | p.Q541* | c.1621C>T | COSV65038659 | Tier2 | 1 / 117 |
| chrX:44922952 | G | GGAAAC | 16 / 29 | p.V607fs | c.1815_1819dupAAACG | Tier2 | 1 / 117 | |
| chrX:44923035 | G | A | 16 / 29 | p.W632* | c.1896G>A | COSV65037694 | Tier2 | 1 / 117 |
| chrX:44923042 | C | T | 16 / 29 | p.Q635* | c.1903C>T | Tier2 | 1 / 117 | |
| chrX:44928869 | G | T | 17 / 29 | p.E657* | c.1969G>T | COSV65041414 | Tier2 | 1 / 117 |
| chrX:44928872 | C | T | 17 / 29 | p.R658* | c.1972C>T | COSV65038721 | Tier2 | 2 / 117 |
| chrX:44928875 | CCT | C | 17 / 29 | p.S661fs | c.1982_1983delCT | COSV100937867 | Tier2 | 1 / 117 |
| chrX:44929133 | G | T | 17 / 29 | p.E745* | c.2233G>T | Tier2 | 1 / 117 | |
| chrX:44929237 | GCT | G | 17 / 29 | p.S781fs | c.2342_2343delCT | COSV65042387 | Tier2 | 1 / 117 |
| chrX:44929411 | CACAA | C | 17 / 29 | p.N839fs | c.2515_2518delAACA | COSV65039572 | Tier2 | 1 / 117 |
| chrX:44938435 | G | T | 20 / 29 | p.E995* | c.2983G>T | COSV65043367 | Tier2 | 1 / 117 |
| chrX:44938447 | G | T | 20 / 29 | p.E999* | c.2995G>T | COSV65038034 | Tier2 | 1 / 117 |
| chrX:44945187 | G | T | 24 / 29 | p.E1171* | c.3511G>T | Tier2 | 1 / 117 | |
| chrX:44949076 | C | T | 25 / 29 | p.R1213* | c.3637C>T | COSV65038395 | Tier2 | 2 / 117 |
| chrX:44966716 | G | T | 27 / 29 | p.G1314* | c.3940G>T | COSV65040483 | Tier2 | 1 / 117 |
| chrX:44966750 | AAGAACCAGCTCATT | A | 27 / 29 | p.E1326fs | c.3976_3989delGAACCAGCTCATTA | Tier2 | 1 / 117 | |
| chrX:44969327 | G | T | 28 / 29 | p.E1337* | c.4009G>T | COSV65042113 | Tier2 | 1 / 117 |
| chrX:44969369 | C | T | 28 / 29 | p.R1351* | c.4051C>T | COSV65038966 | Tier2 | 1 / 117 |
| chrX:44969405 | C | T | 28 / 29 | p.R1363* | c.4087C>T | COSV100938725 | Tier2 | 1 / 117 |

