Side Menu
MSH6
Basic information
- Official gene symbol
-
mutS homolog 6
- Conventional gene symbol
-
GTBP, GTMBP, HNPCC5, HSAP, p160
- Classification
-
Tumor suppressor gene
- Official gene ID
- Transcript ID
- Functional classification
-
Genome maintenance
- Signaling pathway
-
Core DNA Damage Response
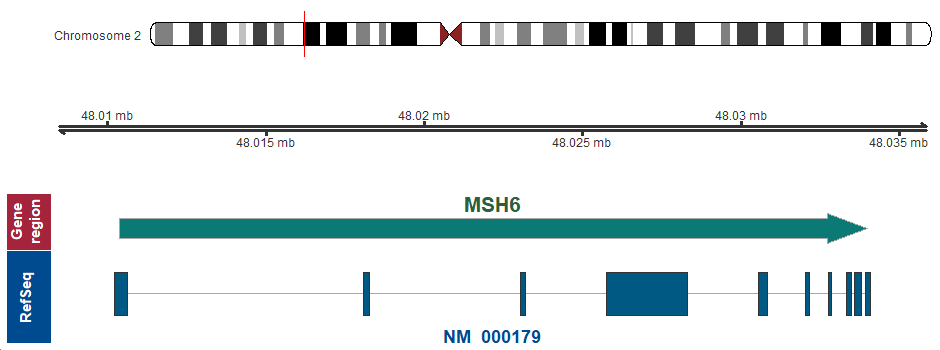
- Chromosomal location
-
2p16.3 (chr2:48010373..48033999)
- Protein length
-
1,360
Chromosome ideogram
Frequency of somatic alterationsTumor type
Base substitutions, insertions, and deletions

Copy number alterations

Distribution of tumor mutation burdenTMB

List of driver somatic mutations
| Genomic Coordinates (GRCh37/hg19) | Reference | Variant | Exon | Amino Acid change | Coding DNA change | COSMIC (v92) | Classification | No. of Samples |
|---|---|---|---|---|---|---|---|---|
| chr2:48025798 | G | T | 4 / 10 | p.E226* | c.676G>T | COSV52283418 | Tier2 | 1 / 93 |
| chr2:48025840 | C | T | 4 / 10 | p.R240* | c.718C>T | COSV52275939 | Tier2 | 1 / 93 |
| chr2:48025877 | C | G | 4 / 10 | p.S252* | c.755C>G | COSV99315000 | Tier2 | 1 / 93 |
| chr2:48026114 | C | G | 4 / 10 | p.S331* | c.992C>G | Tier2 | 1 / 93 | |
| chr2:48026204 | G | A | 4 / 10 | p.R361H | c.1082G>A | COSV52277030 | Tier2 | 2 / 93 |
| chr2:48026217 | G | A | 4 / 10 | p.W365* | c.1095G>A | Tier2 | 2 / 93 | |
| chr2:48026756 | AAG | A | 4 / 10 | p.E546fs | c.1637_1638delAG | Tier2 | 1 / 93 | |
| chr2:48027493 | C | CGTCT | 4 / 10 | p.D793fs | c.2372_2375dupGTCT | Tier2 | 1 / 93 | |
| chr2:48027541 | G | T | 4 / 10 | p.E807* | c.2419G>T | Tier2 | 1 / 93 | |
| chr2:48027661 | G | T | 4 / 10 | p.E847* | c.2539G>T | COSV52290329 | Tier2 | 1 / 93 |
| chr2:48027751 | G | T | 4 / 10 | p.E877* | c.2629G>T | COSV52286026 | Tier2 | 1 / 93 |
| chr2:48027844 | G | T | 4 / 10 | p.E908* | c.2722G>T | COSV52277227 | Tier2 | 1 / 93 |
| chr2:48027853 | C | T | 4 / 10 | p.R911* | c.2731C>T | COSV52274832 | Tier2 | 1 / 93 |
| chr2:48027900 | TA | T | 4 / 10 | p.I927fs | c.2779delA | Tier2 | 1 / 93 | |
| chr2:48027949 | GACATAAGAGAA | G | 4 / 10 | p.D943fs | c.2829_2839delCATAAGAGAAA | Tier2 | 1 / 93 | |
| chr2:48027999 | C | CAACA | 4 / 10 | p.R961fs | c.2878_2881dupAACA | Tier2 | 1 / 93 | |
| chr2:48028049 | G | A | 4 / 10 | p.R976H | c.2927G>A | COSV52275309 | Tier2 | 1 / 93 |
| chr2:48028142 | G | A | 4 / 10 | p.W1007* | c.3020G>A | Tier2 | 1 / 93 | |
| chr2:48028219 | AT | A | 4 / 10 | p.M1033fs | c.3098delT | Tier2 | 1 / 93 | |
| chr2:48028225 | C | T | 4 / 10 | p.R1035* | c.3103C>T | COSV52277143 | Tier2 | 1 / 93 |
| chr2:48030571 | G | A | 5 / 10 | p.C1062Y | c.3185G>A | Tier2 | 1 / 93 | |
| chr2:48030820 | G | GA | 5 / 10 | p.Q1146fs | c.3435dupA | Tier2 | 1 / 93 | |
| chr2:48033396 | G | T | 8 / 10 | p.E1234* | c.3700G>T | COSV52275668 | Tier2 | 2 / 93 |
| chr2:48033589 | AG | A | 9 / 10 | p.A1268fs | c.3802delG | Tier2 | 1 / 93 | |
| chr2:48033753 | G | T | 9 / 10 | p.E1322* | c.3964G>T | COSV52274101 | Tier2 | 2 / 93 |

