Side Menu
NOTCH1
Basic information
- Official gene symbol
-
notch receptor 1
- Conventional gene symbol
-
AOS5, AOVD1, TAN1, hN1
- Classification
-
Oncogene / Tumor suppressor gene
- Official gene ID
- Transcript ID
- Functional classification
-
Differentiation
- Signaling pathway
-
NOTCH
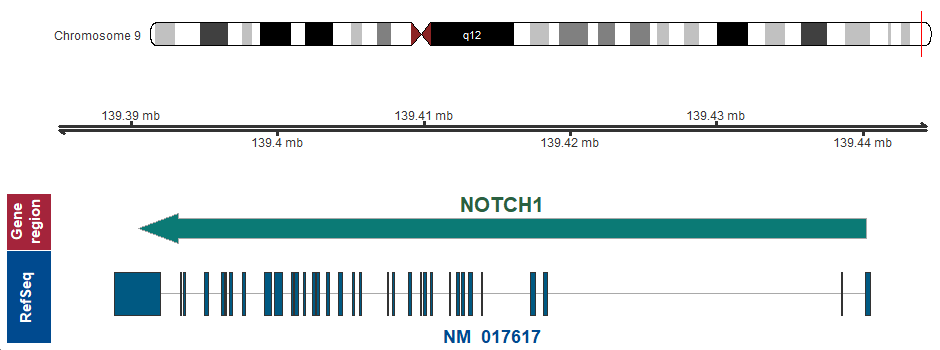
- Chromosomal location
-
9q34.3 (chr9:139390523..139440238, complement)
- Protein length
-
2,555
Chromosome ideogram
Frequency of somatic alterationsTumor type
Base substitutions, insertions, and deletions

Copy number alterations

Distribution of tumor mutation burdenTMB

List of driver somatic mutations
| Genomic Coordinates (GRCh37/hg19) | Reference | Variant | Exon | Amino Acid change | Coding DNA change | COSMIC (v92) | Classification | No. of Samples |
|---|---|---|---|---|---|---|---|---|
| chr9:139438540 | G | A | 2 / 34 | p.Q26* | c.76C>T | Tier2 | 1 / 272 | |
| chr9:139418339 | TCCACCACGTGGCATGTCCCGGCGTTCTTGCA | T | 3 / 34 | p.C68fs | c.202_232delTGCAAGAACGCCGGGACATGCCACGTGGTGG | Tier2 | 1 / 272 | |
| chr9:139418400 | G | A | 3 / 34 | p.Q58* | c.172C>T | Tier2 | 1 / 272 | |
| chr9:139417477 | G | T | 4 / 34 | p.C189* | c.567C>A | Tier2 | 1 / 272 | |
| chr9:139417556 | ATG | A | 4 / 34 | p.I163fs | c.486_487delCA | Tier2 | 1 / 272 | |
| chr9:139417634 | G | T | 4 / 34 | p.S137* | c.410C>A | Tier2 | 1 / 272 | |
| chr9:139413923 | G | T | 5 / 34 | p.Y279* | c.837C>A | Tier2 | 1 / 272 | |
| chr9:139413997 | C | A | 5 / 34 | p.E255* | c.763G>T | Tier2 | 1 / 272 | |
| chr9:139412321 | G | A | 8 / 34 | p.Q442* | c.1324C>T | COSV53049863 | Tier2 | 1 / 272 |
| chr9:139411805 | C | CT | 9 / 34 | p.D492fs | c.1473dupA | Tier2 | 1 / 272 | |
| chr9:139410452 | G | GT | 10 / 34 | p.Y550fs | c.1649dupA | COSV53025560 | Tier2 | 1 / 272 |
| chr9:139410455 | A | AGT | 10 / 34 | p.Y550fs | c.1645_1646dupAC | Tier2 | 1 / 272 | |
| chr9:139410521 | G | C | 10 / 34 | p.Y527* | c.1581C>G | Tier2 | 1 / 272 | |
| chr9:139409060 | GC | G | 13 / 34 | p.R703fs | c.2108delG | Tier2 | 1 / 272 | |
| chr9:139409077 | TGATGCCGTCCTCGCAG | T | 13 / 34 | p.C693fs | c.2076_2091delCTGCGAGGACGGCATC | Tier2 | 1 / 272 | |
| chr9:139407925 | C | A | 14 / 34 | p.E758* | c.2272G>T | Tier2 | 1 / 272 | |
| chr9:139407553 | GCA | G | 15 / 34 | p.A796fs | c.2385_2386delTG | Tier2 | 1 / 272 | |
| chr9:139405664 | C | A | 16 / 34 | p.E843* | c.2527G>T | COSV53039854 | Tier2 | 1 / 272 |
| chr9:139405159 | G | A | 17 / 34 | p.Q896* | c.2686C>T | COSV53102866 | Tier2 | 1 / 272 |
| chr9:139405192 | G | A | 17 / 34 | p.Q885* | c.2653C>T | COSV53099073 | Tier2 | 1 / 272 |
| chr9:139404349 | G | GT | 18 / 34 | p.G936fs | c.2804_2805insA | Tier2 | 1 / 272 | |
| chr9:139403380 | T | TCCTGACAGGTG | 19 / 34 | p.D1038fs | c.3102_3112dupCACCTGTCAGG | Tier2 | 1 / 272 | |
| chr9:139402775 | G | GTGGGTCTGCCAGCATTTGC | 20 / 34 | p.H1078fs | c.3215_3233dupGCAAATGCTGGCAGACCCA | Tier2 | 1 / 272 | |
| chr9:139402785 | C | T | 20 / 34 | p.W1075* | c.3224G>A | COSV99493676 | Tier2 | 1 / 272 |
| chr9:139402807 | AGGGC | A | 20 / 34 | p.P1067fs | c.3198_3201delGCCC | Tier2 | 1 / 272 | |
| chr9:139402567 | T | TG | 21 / 34 | p.Q1117fs | c.3349dupC | Tier2 | 1 / 272 | |
| chr9:139401760 | G | A | 22 / 34 | p.Q1214* | c.3640C>T | COSV53031275 | Tier2 | 1 / 272 |
| chr9:139401256 | G | T | 23 / 34 | p.C1271* | c.3813C>A | Tier2 | 1 / 272 | |
| chr9:139401261 | C | A | 23 / 34 | p.E1270* | c.3808G>T | Tier2 | 1 / 272 | |
| chr9:139400978 | CCG | C | 24 / 34 | p.A1338fs | c.4013_4014delCG | Tier2 | 1 / 272 | |
| chr9:139399888 | C | T | 25 / 34 | p.W1487* | c.4460G>A | COSV53031807 | Tier2 | 1 / 272 |
| chr9:139399191 | G | T | 26 / 34 | p.S1651* | c.4952C>A | Tier2 | 1 / 272 | |
| chr9:139399237 | C | A | 26 / 34 | p.E1636* | c.4906G>T | Tier2 | 1 / 272 | |
| chr9:139399456 | C | A | 26 / 34 | p.E1563* | c.4687G>T | Tier2 | 1 / 272 | |
| chr9:139399481 | G | T | 26 / 34 | p.C1554* | c.4662C>A | Tier2 | 1 / 272 | |
| chr9:139397646 | C | A | 27 / 34 | p.E1719* | c.5155G>T | COSV99490401 | Tier2 | 1 / 272 |
| chr9:139396859 | ACGAAGAACAGAAG | A | 28 / 34 | p.L1746fs | c.5236_5248delCTTCTGTTCTTCG | Tier2 | 1 / 272 | |
| chr9:139396479 | C | A | 29 / 34 | p.E1816* | c.5446G>T | Tier2 | 1 / 272 | |
| chr9:139396275 | TGC | T | 30 / 34 | p.R1854fs | c.5561_5562delGC | Tier2 | 1 / 272 | |
| chr9:139395053 | CG | C | 31 / 34 | p.R1962fs | c.5884delC | Tier2 | 1 / 272 | |
| chr9:139393596 | G | C | 32 / 34 | p.S2017* | c.6050C>G | COSV53042787 | Tier2 | 1 / 272 |
| chr9:139393696 | G | A | 32 / 34 | p.R1984* | c.5950C>T | COSV53039810 | Tier2 | 2 / 272 |

