Side Menu
RBM10
Basic information
- Official gene symbol
-
RNA binding motif protein 10
- Conventional gene symbol
-
DXS8237E, GPATC9, GPATCH9, S1-1, TARPS, ZRANB5
- Classification
-
Tumor suppressor gene
- Official gene ID
- Transcript ID
- Functional classification
-
Transcriptional regulation
- Signaling pathway
-
RNA metabolism
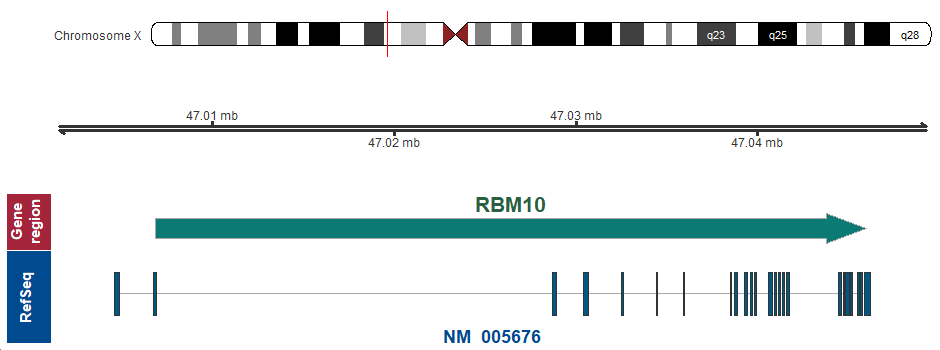
- Chromosomal location
-
Xp11.3 (chrX:47006881..47045998)
- Protein length
-
930
Chromosome ideogram
Frequency of somatic alterationsTumor type
Base substitutions, insertions, and deletions

Copy number alterations

Distribution of tumor mutation burdenTMB

List of driver somatic mutations
| Genomic Coordinates (GRCh37/hg19) | Reference | Variant | Exon | Amino Acid change | Coding DNA change | COSMIC (v92) | Classification | No. of Samples |
|---|---|---|---|---|---|---|---|---|
| chrX:47028763 | C | T | 3 / 24 | p.Q23* | c.67C>T | Tier2 | 1 / 127 | |
| chrX:47028790 | C | CGA | 3 / 24 | p.D33fs | c.97_98dupGA | Tier2 | 1 / 127 | |
| chrX:47028828 | T | G | 3 / 24 | p.Y44* | c.132T>G | COSV61309029 | Tier2 | 1 / 127 |
| chrX:47035912 | TC | T | 7 / 24 | p.L198fs | c.592delC | Tier2 | 1 / 127 | |
| chrX:47035983 | A | T | 7 / 24 | p.K221* | c.661A>T | Tier2 | 1 / 127 | |
| chrX:47038526 | C | T | 8 / 24 | p.R230* | c.688C>T | COSV61309636 | Tier2 | 2 / 127 |
| chrX:47038726 | C | T | 9 / 24 | p.Q245* | c.733C>T | COSV61313432 | Tier2 | 1 / 127 |
| chrX:47038751 | TG | T | 9 / 24 | p.D254fs | c.760delG | Tier2 | 1 / 127 | |
| chrX:47038846 | C | T | 9 / 24 | p.Q285* | c.853C>T | COSV61312003 | Tier2 | 1 / 127 |
| chrX:47039406 | CG | C | 10 / 24 | p.G344fs | c.1031delG | Tier2 | 1 / 127 | |
| chrX:47039613 | GGCAGCCCA | G | 11 / 24 | p.Q358fs | c.1068_1075delAGCCCAGC | Tier2 | 1 / 127 | |
| chrX:47040939 | CTG | C | 14 / 24 | p.S492fs | c.1473_1474delGT | Tier2 | 1 / 127 | |
| chrX:47041199 | ACC | A | 15 / 24 | p.H544fs | c.1629_1630delCC | Tier2 | 1 / 127 | |
| chrX:47041211 | TCTGCTCTCCCACCGGCTACCAGCCCCACTGCCCAGGAATC | T | 15 / 24 | p.A548fs | c.1642_1681delGCTCTCCCACCGGCTACCAGCCCCACTGCCCAGGAATCCT | Tier2 | 1 / 127 | |
| chrX:47041384 | GA | G | 16 / 24 | p.T577fs | c.1729delA | Tier2 | 1 / 127 | |
| chrX:47041599 | G | A | 17 / 24 | p.W608* | c.1824G>A | Tier2 | 1 / 127 | |
| chrX:47044550 | G | T | 18 / 24 | p.E683* | c.2047G>T | COSV61308334 | Tier2 | 1 / 127 |
| chrX:47044595 | G | T | 18 / 24 | p.E698* | c.2092G>T | COSV61307701 | Tier2 | 1 / 127 |
| chrX:47044713 | G | T | 19 / 24 | p.E705* | c.2113G>T | COSV61307871 | Tier2 | 1 / 127 |
| chrX:47044753 | GT | G | 19 / 24 | p.S718fs | c.2154delT | Tier2 | 1 / 127 | |
| chrX:47044892 | G | T | 20 / 24 | p.E740* | c.2218G>T | COSV61309228 | Tier2 | 1 / 127 |
| chrX:47044946 | A | T | 20 / 24 | p.K758* | c.2272A>T | Tier2 | 1 / 127 | |
| chrX:47045124 | G | T | 21 / 24 | p.E789* | c.2365G>T | COSV61309684 | Tier2 | 1 / 127 |
| chrX:47045149 | C | G | 21 / 24 | p.S797* | c.2390C>G | Tier2 | 1 / 127 | |
| chrX:47045464 | C | T | 22 / 24 | p.Q811* | c.2431C>T | Tier2 | 1 / 127 | |
| chrX:47045518 | G | T | 22 / 24 | p.E829* | c.2485G>T | COSV61311914 | Tier2 | 1 / 127 |
| chrX:47045678 | GGACGGGCTGGGCA | G | 23 / 24 | p.D854fs | c.2561_2573delACGGGCTGGGCAG | Tier2 | 1 / 127 |

