Side Menu
SMAD4
Basic information
- Official gene symbol
-
SMAD family member 4
- Conventional gene symbol
-
DPC4, JIP, MADH4, MYHRS
- Classification
-
Tumor suppressor gene
- Official gene ID
- Transcript ID
- Functional classification
-
Tumor growth and progression
- Signaling pathway
-
TGF-B
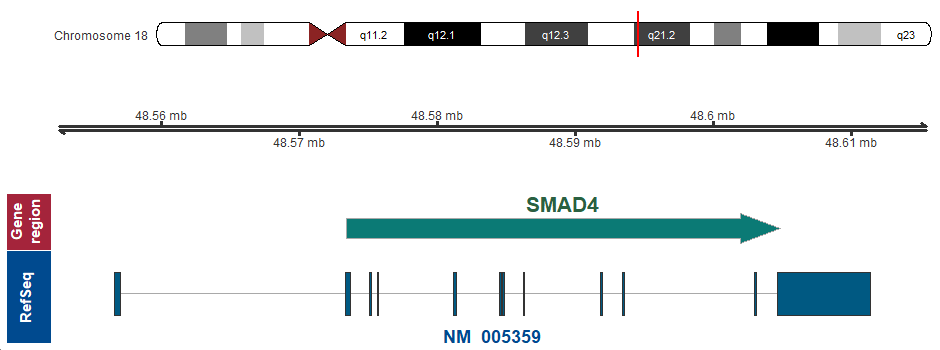
- Chromosomal location
-
18q21.2 (chr18:48573417..48604837)
- Protein length
-
552
Chromosome ideogram
Pathway map

Frequency of somatic alterationsTumor type
Base substitutions, insertions, and deletions

Copy number alterations

Distribution of tumor mutation burdenTMB

List of driver somatic mutations
| Genomic Coordinates (GRCh37/hg19) | Reference | Variant | Exon | Amino Acid change | Coding DNA change | COSMIC (v92) | Classification | No. of Samples |
|---|---|---|---|---|---|---|---|---|
| chr18:48573504 | GGA | G | 2 / 12 | p.S32fs | c.94_95delAG | COSV61685245 | Tier2 | 3 / 185 |
| chr18:48573507 | G | T | 2 / 12 | p.E31* | c.91G>T | Tier2 | 1 / 185 | |
| chr18:48573573 | G | T | 2 / 12 | p.E53* | c.157G>T | COSV61691604 | Tier2 | 1 / 185 |
| chr18:48573639 | C | T | 2 / 12 | p.Q75* | c.223C>T | Tier2 | 1 / 185 | |
| chr18:48575111 | C | T | 3 / 12 | p.P102L | c.305C>T | COSV61696589 | Tier2 | 1 / 185 |
| chr18:48575206 | G | T | 3 / 12 | p.E134* | c.400G>T | Tier2 | 1 / 185 | |
| chr18:48575671 | C | G | 4 / 12 | p.S144* | c.431C>G | COSV61685296 | Tier2 | 2 / 185 |
| chr18:48581253 | C | CA | 5 / 12 | p.S187fs | c.559dupA | Tier2 | 1 / 185 | |
| chr18:48581335 | C | CTT | 5 / 12 | p.P215fs | c.641_642dupTT | Tier2 | 1 / 185 | |
| chr18:48584499 | GC | G | 6 / 12 | p.P225fs | c.674delC | Tier2 | 1 / 185 | |
| chr18:48584540 | T | A | 6 / 12 | p.L238* | c.713T>A | Tier2 | 1 / 185 | |
| chr18:48584560 | C | T | 6 / 12 | p.Q245* | c.733C>T | COSV61684971 | Tier2 | 1 / 185 |
| chr18:48584569 | C | T | 6 / 12 | p.Q248* | c.742C>T | COSV61689992 | Tier2 | 1 / 185 |
| chr18:48584593 | C | T | 6 / 12 | p.Q256* | c.766C>T | COSV61686540 | Tier2 | 1 / 185 |
| chr18:48584607 | C | G | 6 / 12 | p.Y260* | c.780C>G | Tier2 | 1 / 185 | |
| chr18:48584722 | CCTGGA | C | 7 / 12 | p.T269fs | c.806_810delCTGGA | Tier2 | 1 / 185 | |
| chr18:48586232 | T | TTAGG | 8 / 12 | p.W302fs | c.905-3_905dupTAGG | Tier2 | 1 / 185 | |
| chr18:48586262 | C | T | 8 / 12 | p.Q311* | c.931C>T | COSV61687817 | Tier2 | 1 / 185 |
| chr18:48591806 | G | A | 9 / 12 | p.W323* | c.969G>A | Tier2 | 1 / 185 | |
| chr18:48591808 | G | A | 9 / 12 | p.C324Y | c.971G>A | COSV61695700 | Tier2 | 1 / 185 |
| chr18:48591816 | GCTTA | G | 9 / 12 | p.Y328fs | c.983_986delACTT | Tier2 | 1 / 185 | |
| chr18:48591825 | G | T | 9 / 12 | p.E330* | c.988G>T | COSV61684539 | Tier2 | 1 / 185 |
| chr18:48591826 | A | C | 9 / 12 | p.E330A | c.989A>C | COSV61694916 | Tier1 | 1 / 185 |
| chr18:48591837 | C | T | 9 / 12 | p.Q334* | c.1000C>T | COSV61687277 | Tier2 | 1 / 185 |
| chr18:48591888 | G | C | 9 / 12 | p.D351H | c.1051G>C | COSV61684645 | Tier1 | 1 / 185 |
| chr18:48591918 | C | T | 9 / 12 | p.R361C | c.1081C>T | COSV61683972 | Tier1 | 7 / 185 |
| chr18:48591918 | C | G | 9 / 12 | p.R361G | c.1081C>G | COSV61686083 | Tier2 | 4 / 185 |
| chr18:48591918 | C | A | 9 / 12 | p.R361S | c.1081C>A | COSV61685418 | Tier1 | 2 / 185 |
| chr18:48591919 | G | A | 9 / 12 | p.R361H | c.1082G>A | COSV61684056 | Tier1 | 21 / 185 |
| chr18:48591947 | C | CG | 9 / 12 | p.H371fs | c.1110_1111insG | Tier2 | 1 / 185 | |
| chr18:48593405 | G | T | 10 / 12 | p.G386C | c.1156G>T | COSV61697840 | Tier2 | 1 / 185 |
| chr18:48593406 | G | T | 10 / 12 | p.G386V | c.1157G>T | COSV61688134 | Tier2 | 1 / 185 |
| chr18:48593505 | G | T | 10 / 12 | p.G419V | c.1256G>T | COSV61685351 | Tier2 | 1 / 185 |
| chr18:48604656 | A | C | 12 / 12 | p.D493A | c.1478A>C | COSV61689238 | Tier2 | 1 / 185 |
| chr18:48604665 | G | A | 12 / 12 | p.R496H | c.1487G>A | COSV61684216 | Tier2 | 5 / 185 |
| chr18:48604668 | G | A | 12 / 12 | p.R497H | c.1490G>A | COSV61687877 | Tier2 | 4 / 185 |
| chr18:48604705 | G | A | 12 / 12 | p.W509* | c.1527G>A | COSV61689637 | Tier2 | 1 / 185 |
| chr18:48604736 | G | T | 12 / 12 | p.E520* | c.1558G>T | COSV61685467 | Tier2 | 1 / 185 |
| chr18:48604747 | C | A | 12 / 12 | p.C523* | c.1569C>A | COSV100747628 | Tier2 | 1 / 185 |
| chr18:48604764 | T | TA | 12 / 12 | p.H530fs | c.1587dupA | COSV61687828 | Tier2 | 1 / 185 |
| chr18:48604778 | C | T | 12 / 12 | p.Q534* | c.1600C>T | COSV61688543 | Tier2 | 1 / 185 |
| chr18:48604787 | G | T | 12 / 12 | p.D537Y | c.1609G>T | COSV61684436 | Tier1 | 1 / 185 |
| chr18:48604788 | A | G | 12 / 12 | p.D537G | c.1610A>G | COSV61685387 | Tier2 | 4 / 185 |
| chr18:48604789 | C | A | 12 / 12 | p.D537E | c.1611C>A | COSV61687202 | Tier2 | 1 / 185 |
| chr18:48604790 | G | T | 12 / 12 | p.E538* | c.1612G>T | COSV61684525 | Tier2 | 2 / 185 |

