Announcement: Release plan for the English version of the 2022 JCGA database
The next-generation sequencing (NGS)-based cancer gene panel tests have been used in clinical practice, in Japan, since June 2019. A cancer genome database of Japanese patients, with mutational profiles unaffected by racial differences, has been extremely important in improving the interpretation of the detected gene alterations. Thus, we constructed the Japanese version of the Cancer Genome Atlas (JCGA) based on the whole-exome sequencing data of 4,907 surgically resected primary tumor samples obtained from 4,753 Japanese patients with cancer who were enrolled in Project HOPE*. Project HOPE is a prospective molecular profiling study for multiple tumor types using multi-omics technology utilizing NGS. It was launched at the Shizuoka Cancer Center, in January 2014. The cohort consisted of 134 tumor types classified based on the criteria set by Oncotree and/or The Cancer Genome Atlas.
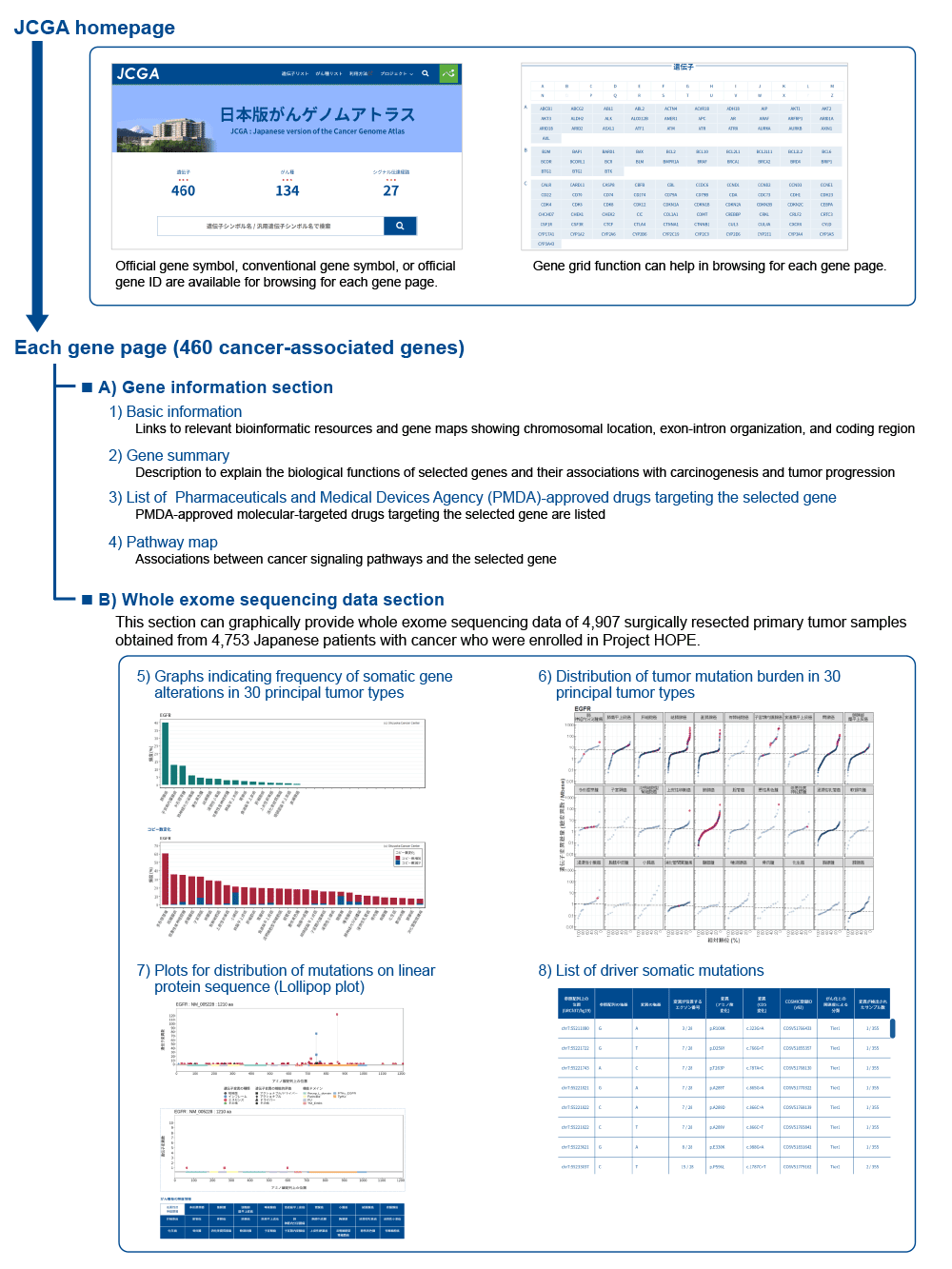
The JCGA graphically provides genome information on 460 cancer-associated genes, including the 336 genes that are involved in two NGS-based cancer gene panel tests approved by the Pharmaceuticals and Medical Devices Agency (PMDA). The JCGA has been used by the molecular tumor boards that are the so-called expert panel of our institute to interpret the gene alterations detected in the NGS-based cancer gene panel tests and to prepare the reports that explain putative biological causes that drive tumor progression. In the current version of the JCGA, most of the contents are written in Japanese; this aids physicians explain the results of NGS-based cancer gene panel tests and enables patients and their families to obtain further information regarding the detected gene alterations.
We are currently constructing an English version of the JCGA, which will be released in 2022. It will provide cancer genome information of Japanese patients to cancer researchers worldwide and will accelerate the evaluation of differences among ethnic groups in the distribution of histological types and genomic signature.
September 1, 2021

- *Biomed Res. 2014; 35: 407-412. (PMID: 25743347)
- *Cancer Sci. 2020; 111: 687-699. (PMID: 31863614)
Overview of the JCGA database
On the homepage of the JCGA, users can search by entering the official gene symbol, trivial gene symbol, or official gene ID (Entrez ID) in the search window or gene grid function. The resulting display page of each gene provides following eight contents: 1) basic information, such as links to relevant bioinformatic resources and gene maps showing chromosomal location, exon-intron organization, and coding region; 2) gene summary, which explains the biological function and associations with carcinogenesis and tumor progression; 3) list of PMDA-approved drugs targeting the selected gene; 4) pathway map, which shows the association between cancer signaling pathways and the selected gene; 5) graphs, which indicate the frequency of somatic gene alterations in 30 principal tumor types; 6) distribution of the tumor mutational burden (TMB) in 30 principle tumor types sorted in ascending order of the median value of TMB in each tumor type; 7) plots for the distribution of mutations in the linear protein sequence, which are the so-called lollipop plots; and 8) list of driver somatic mutations. These contents are divided into two sections: A) gene information section (contents 1-4), which provides biological background for each gene, and B) whole exome sequencing data section (contents 5-8), which graphically provides whole exome sequencing data of 4,907 primary tumor samples of Project HOPE.
The frequency of somatic gene alterations (content 5) are useful in estimating the primary tumor types in patient with unknown primary tumors. The TMB distribution (content 6) can be used to assess its relative ranking of TMB of patients, and is useful in predicting the therapeutic effect of immune checkpoint inhibitors. The “comparison of lollipop plots” function in content 7 can be used to compare the distribution of the detected mutations in the linear protein sequence between all tumor types and the selected tumor type and can contribute to the identification of tumor types that frequently harbor mutations located in hotspot or functional domains.